Softwares and Web-tools
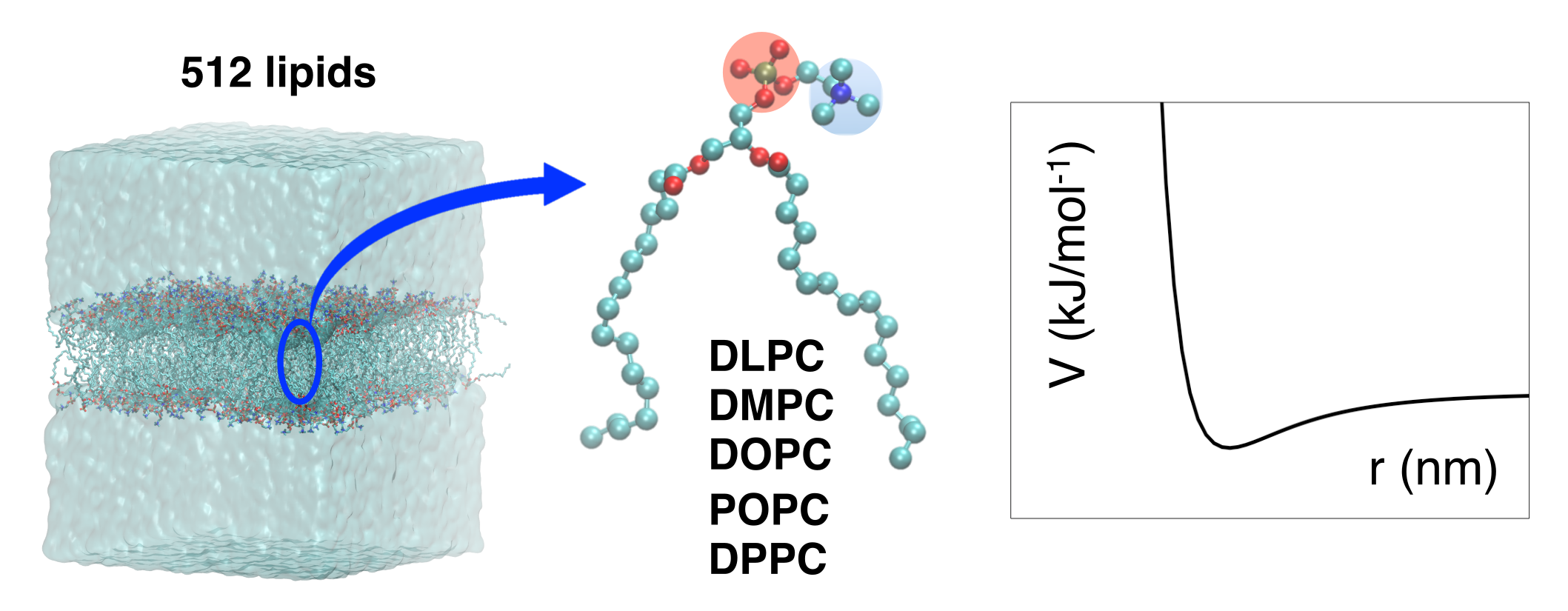
Lipid parameters for Molecular Dynamics simulations of biomembrane have been updated to the most recent version of the GROMOS 54a8 force field to improve solvation properties in the head region. The resulting parameters and topology files can be found here.
UNIPPIN is an integrated protein-protein interaction network the group has been collecting and curating for our analyses on protein-protein interactions. Find out more here.
BRepertoire is a suite of user-friendly web-based software tools for large-scale statistical analyses of antibody repertoire data. It works on data preprocessed by IMGT, and performs statistical and comparative analyses on-the-fly with versatile plotting options.
Cite: Margreitter C, Lu HC, Townsend C, Stewart A, Dunn-Walters DK, Fraternali F BRepertoire: a user-friendly web server for analysing antibody repertoire data, Nucleic Acids Research, gky276, https://doi.org/10.1093/nar/gky276
TITINdb is a web application which integrates titin structure, sequence, isoform, variant and disease information. Disease-associated nonsynonymous SNPs (nsSNPs) from the literature and population nsSNPs from the 1000 genomes project and ExAC database to be visualised on structure. Additionally the impact of these nsSNPs has been predicted computationally using both structure and sequence based methods.
Cite: Laddach A, Gautel M, Fraternali F TITINdb: a Computational Tool to Assess Titin’s Role as a Disease Gene. Bioinformatics 2017 btx424. doi: 10.1093/bioinformatics/btx424, PMID: [29077808](http://www.ncbi.nlm.nih.gov/pubmed/29077808
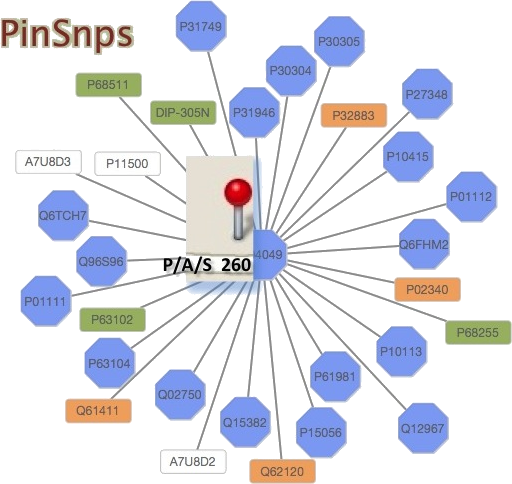
PinSnps is an interactive tool for the analysis of common variants, disease-related single-nucleotide polymorphisms (SNPs), Post-Translational Modifications (PTMs) and related functional sites mapped onto human protein interaction networks. One can search for and retrieve information about SNPs data and interacting proteins of the query.
Cite: Lu, HC, Herrera Braga, J, Fraternali, F. PinSnps: structural and functional analysis of SNPs in the context of protein interaction networks. Bioinformatics 32(16):2534-6, 2016. PMID: 27153707
POPSCOMP is a method to analyse interactions between individual complex components of proteins and/or nucleic acids by calculating the solvent accessible surface area (SASA) buried upon complex formation. It is based on POPS, which calculates SASAs of individual proteins and nucleic acid molecules at atomic (default) and residue (coarse-grained) level.
The underlying heuristic algorithm of POPS uses a geometric approximation to the shielding of atomic surface areas by nearest neighbour atoms.
Cite:
- Kleinjung, J, Fraternali, F. POPSCOMP: An automated interaction analysis of biomolecular complexes. Nuc. Acids Res. 33:W342-W346, 2005 PMID: 15980485
- Cavallo, L, Kleinjung, J, Fraternali, F. POPS: a fast algorithm for Solvent Accessible Surface Areas at atomic and residue level. Nuc. Acids Res. 31:3364-3366, 2003 PMID: 12824328