Papers on screening protein-protein interaction inhibitor using crystal lattices
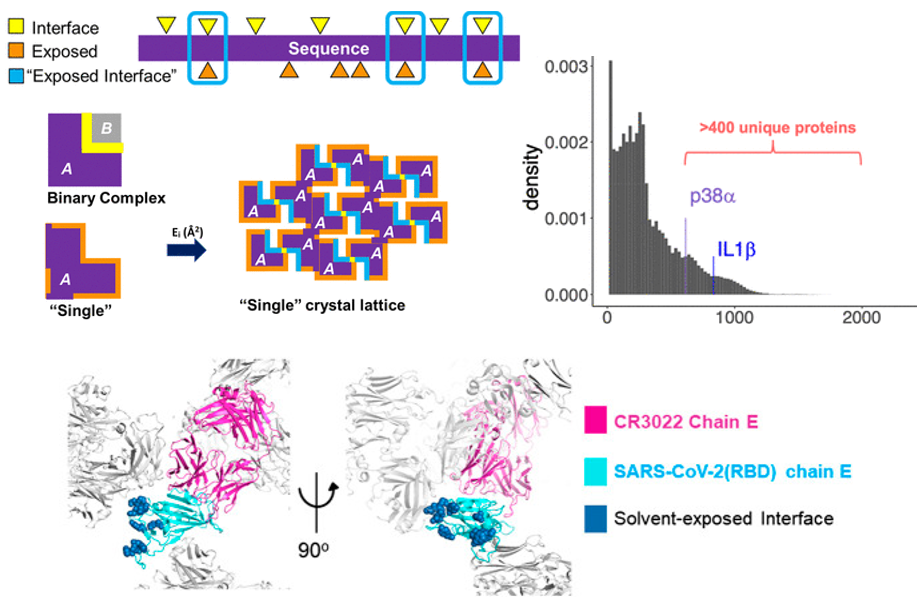
We celebrate two papers on a new in silico method to screen existing crystal lattices available in the Protein Data Bank, for lattices suitable for carrying out fragment soaking experiments to design chemical inhibitors targeting protein-protein interactions! See this paper in J Med Chem for description and application on two attractive therapeutic targets (p38-alpha and interleukin-1-beta).
In a subsequent work now published in Frontiers in Pharmacology we apply the method to analyse existing lattices containing the Receptor Binding Domain (RBD) of the SARS-CoV-2 spike protein. Our collaborator Gian-Felice de Nicola at the Randall has managed to generate a novel crystal form which exposes the interface between RBD and ACE2, making it available for fragment screening to develop inhibitors that target this interaction!